Fitting LoS distributions for HEP#
1. Imports#
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
import seaborn as sns
from fitter import Fitter, get_common_distributions, get_distributions
2. Length-of-stay categories#
Primary Hip
Revision Hip
Primary Knee
Revision Knee
Unicompartmental Primary Knee
Delayed Discharge
3. Read in data#
HipKnee_1320_mffd = pd.read_csv("Test_data/HipKnee_with_los_delay_.csv", usecols = ['admission_date',
'discharge_date',
'spell_los',
'mffd',
'hip_prim_proc',
'knee_prim_proc',
'surgery_site'])
4. Calculate proportions of surgical types within Hips and Knees#
# All hips, all knees
length_hip = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['surgery_site'] == "Site: hip"]
length_knee = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['surgery_site'] == "Site: knee"]
# hip types within all hips
primary_hip_n = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['hip_prim_proc'] == "Primary THR"]
revision_hip_n = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['hip_prim_proc'] == "Revision of hip"]
hip_resurfacing_n = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['hip_prim_proc'] == "Hip resurfacing"]
# knee types within all knees
primary_knee_n = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['knee_prim_proc'] == "Primary TKR"]
revision_knee_n = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['knee_prim_proc'] == "Revision of knee"]
unicompart_knee_n = HipKnee_1320_mffd.loc[HipKnee_1320_mffd['knee_prim_proc'] == "Primary UKR"]
# counts for both delayed and not delayed
print(len(primary_hip_n), len(revision_hip_n), len(hip_resurfacing_n), len(primary_knee_n), len(revision_knee_n), len(unicompart_knee_n))
print(len(primary_hip_n) + len(revision_hip_n) + len(hip_resurfacing_n) + len(primary_knee_n) + len(revision_knee_n) + len(unicompart_knee_n))
3057 482 52 2302 392 679
6964
# prop of hip types within hips
prop_prim_hip = primary_hip_n.shape[0] / length_hip.shape[0]
prop_rev_hip = revision_hip_n.shape[0] / length_hip.shape[0]
prop_hip_resurfacing = hip_resurfacing_n.shape[0] / length_hip.shape[0]
tot_hips = prop_prim_hip + prop_rev_hip + prop_hip_resurfacing
# prop knee types within knees
prop_prim_knee = primary_knee_n.shape[0] / length_knee.shape[0]
prop_rev_knee = revision_knee_n.shape[0] / length_knee.shape[0]
prop_uncompart_knee = unicompart_knee_n.shape[0] / length_knee.shape[0]
tot_knees = prop_prim_knee + prop_rev_knee + prop_uncompart_knee
#proportions for both delayed and not delayed by joint
print(prop_prim_hip, prop_rev_hip, prop_prim_knee, prop_rev_knee, prop_uncompart_knee)
0.8616121758737317 0.1358511837655017 0.6738875878220141 0.11475409836065574 0.1987704918032787
5. Calculate proportions of surgical types within classes: Primary and Revision#
# prop primary types within primaries
prop_primary_hips = primary_hip_n.shape[0] / (primary_hip_n.shape[0] + primary_knee_n.shape[0] + unicompart_knee_n.shape[0])
prop_primary_knees = primary_knee_n.shape[0] / (primary_knee_n.shape[0] + primary_hip_n.shape[0] + unicompart_knee_n.shape[0])
prop_unicompart_knees = unicompart_knee_n.shape[0] / (primary_knee_n.shape[0] + primary_hip_n.shape[0] + unicompart_knee_n.shape[0])
# prop revision types within revisions
prop_revision_hips = revision_hip_n.shape[0] / (revision_hip_n.shape[0] + revision_knee_n.shape[0])
prop_revision_knees = revision_knee_n.shape[0] / (revision_hip_n.shape[0] + revision_knee_n.shape[0])
#proportions for both delayed and not delayed by primary/revision
print(prop_primary_hips, prop_primary_knees, prop_unicompart_knees, prop_revision_hips, prop_revision_knees)
0.5062934746604836 0.3812520702219278 0.1124544551175886 0.551487414187643 0.448512585812357
6. Calculate proportion delayed#
Overall delay for all surgical types
# count of NOT delayed and proportion of delayed
delay_n = HipKnee_1320_mffd[HipKnee_1320_mffd['mffd'].isna()]
delay_prop = (HipKnee_1320_mffd.shape[0] - delay_n.shape[0]) / HipKnee_1320_mffd.shape[0]
delay = HipKnee_1320_mffd.shape[0] - delay_n.shape[0]
delay
529
# overall proportion of patients delayed
delay_prop
0.07596209075244112
# dataframes for checking - patients delayed per surgery
primary_knee_del = primary_knee_n[primary_knee_n['mffd'].notna()]
uni_knee_del = unicompart_knee_n[unicompart_knee_n['mffd'].notna()]
revise_knee_del = revision_knee_n[revision_knee_n['mffd'].notna()]
primary_hip_del = primary_hip_n[primary_hip_n['mffd'].notna()]
revise_hip_del = revision_hip_n[revision_hip_n['mffd'].notna()]
# df patients not delayed
primary_knee = primary_knee_n[primary_knee_n['mffd'].isna()]
uni_knee = unicompart_knee_n[unicompart_knee_n['mffd'].isna()]
revise_knee = revision_knee_n[revision_knee_n['mffd'].isna()]
primary_hip = primary_hip_n[primary_hip_n['mffd'].isna()]
revise_hip = revision_hip_n[revision_hip_n['mffd'].isna()]
# checking proportions per surgery - compare with overall proportion delayed
prop_primary_knee_del = primary_knee_del.shape[0] / (primary_knee.shape[0] + primary_knee_del.shape[0])
prop_revision_knee_del = revise_knee_del.shape[0] / (revise_knee.shape[0] + revise_knee_del.shape[0])
prop_unicom_knee_del = uni_knee_del.shape[0] / (uni_knee.shape[0] + uni_knee_del.shape[0])
prop_primary_hip_del = primary_hip_del.shape[0] / (primary_hip.shape[0] + primary_hip_del.shape[0])
prop_revison_hip_del = revise_hip_del.shape[0] / (revise_hip.shape[0] + revise_hip_del.shape[0])
# proportion of patients delayed per surgery type
print(prop_primary_hip_del, prop_primary_knee_del, prop_revison_hip_del, prop_revision_knee_del, prop_unicom_knee_del)
0.0775269872423945 0.06602953953084274 0.15767634854771784 0.1326530612244898 0.016200294550810016
7. Vectors for fitting no delay LoS#
Save to csv for using empirical distributions in model
primary_knee_v = primary_knee['spell_los'].tolist()
uni_knee_v = uni_knee['spell_los'].tolist()
revise_knee_v = revise_knee['spell_los'].tolist()
primary_hip_v = primary_hip['spell_los'].tolist()
revise_hip_v = revise_hip['spell_los'].tolist()
series_dict = {'Primary Knee': pd.Series(primary_knee_v),
'Unicompart Knee': pd.Series(uni_knee_v),
'Revision Knee': pd.Series(revise_knee_v),
'Primary Hip': pd.Series(primary_hip_v),
'Revision Hip': pd.Series(revise_hip_v)}
#save to df with 0 changed to very small number
df = pd.DataFrame(series_dict)
df.replace(to_replace = 0, value = 0.0001, inplace=True)
df.to_csv('Test_data/los_spells.csv', index=False)
######################################################
#save to df with 0 eliminated
a = [i for i in primary_knee_v if i != 0]
b = [i for i in uni_knee_v if i != 0]
c = [i for i in revise_knee_v if i != 0]
d = [i for i in primary_hip_v if i != 0]
e = [i for i in revise_hip_v if i != 0]
nonzero_dict = {'Primary Knee': pd.Series(a),
'Unicompart Knee': pd.Series(b),
'Revision Knee': pd.Series(c),
'Primary Hip': pd.Series(d),
'Revision Hip': pd.Series(e)}
df2 = pd.DataFrame(nonzero_dict)
df2.to_csv('Test_data/los_spells_no_zero.csv', index=False)
8. Los vector for delayed discharge (all surgical types)#
delayed_los = HipKnee_1320_mffd[HipKnee_1320_mffd['mffd'].notna()]
delayed_los_v = delayed_los['spell_los'].tolist()
9. Visualise LoS distributions#
#plt.hist(primary_knee_v, bins=50);
sns.set_style('white')
sns.set_context("paper", font_scale = 2)
sns.displot(data=primary_knee_v, kind="hist", bins = 50, aspect = 1.5)
<seaborn.axisgrid.FacetGrid at 0x7fe96dbff970>
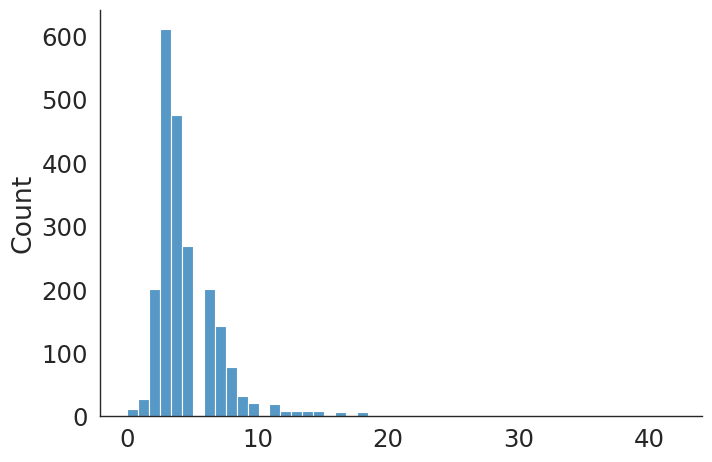
#plt.hist(uni_knee_v, bins=20);
sns.set_style('white')
sns.set_context("paper", font_scale = 2)
sns.displot(data=uni_knee_v, kind="hist", bins = 20, aspect = 1.5)
<seaborn.axisgrid.FacetGrid at 0x7fe965960910>
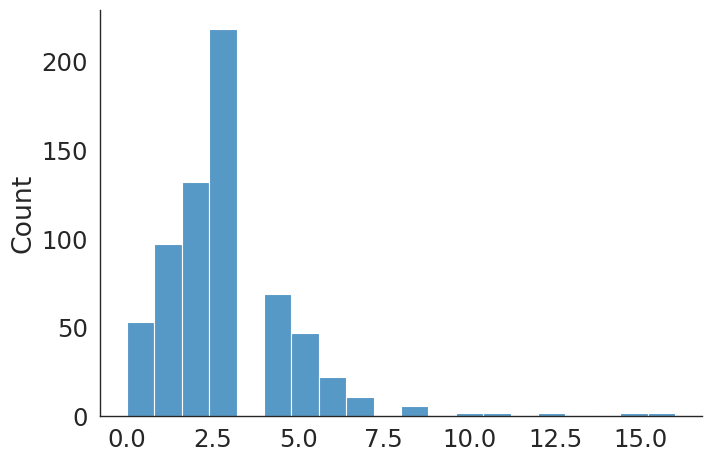
#plt.hist(revise_knee_v, bins=50);
sns.set_style('white')
sns.set_context("paper", font_scale = 2)
sns.displot(data=revise_knee_v, kind="hist", bins = 50, aspect = 1.5)
<seaborn.axisgrid.FacetGrid at 0x7fe96dbffa30>
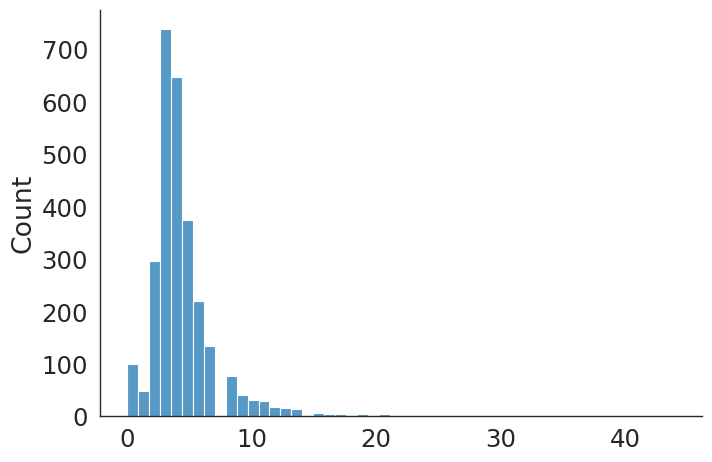
#plt.hist(primary_hip_v, bins=50);
sns.set_style('white')
sns.set_context("paper", font_scale = 2)
sns.displot(data=primary_hip_v, kind="hist", bins = 50, aspect = 1.5)
<seaborn.axisgrid.FacetGrid at 0x7fe9657776a0>
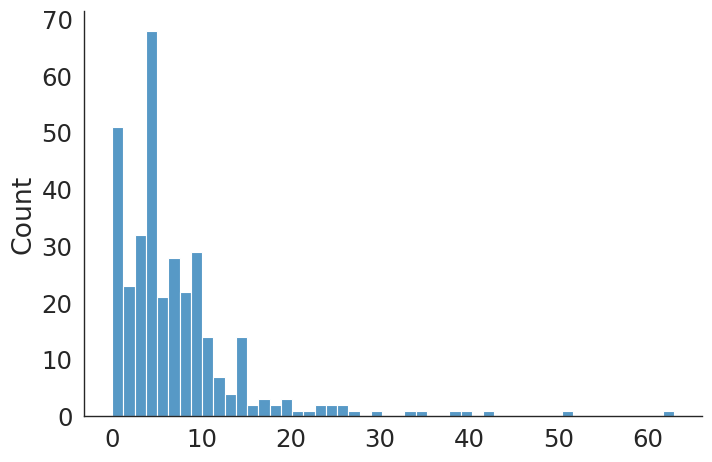
#plt.hist(revise_hip_v, bins=50);
sns.set_style('white')
sns.set_context("paper", font_scale = 2)
sns.displot(revise_hip_v, kind="hist", bins = 50, aspect = 1.5)
<seaborn.axisgrid.FacetGrid at 0x7fe96555e040>
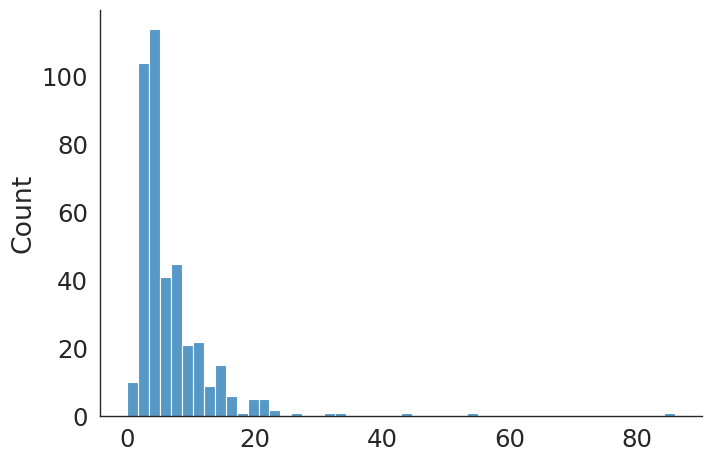
#plt.hist(delayed_los_v, bins=50);
sns.set_style('white')
sns.set_context("paper", font_scale = 2)
sns.displot(delayed_los_v, kind="hist", bins = 50, aspect = 1.5)
<seaborn.axisgrid.FacetGrid at 0x7fe965960cd0>
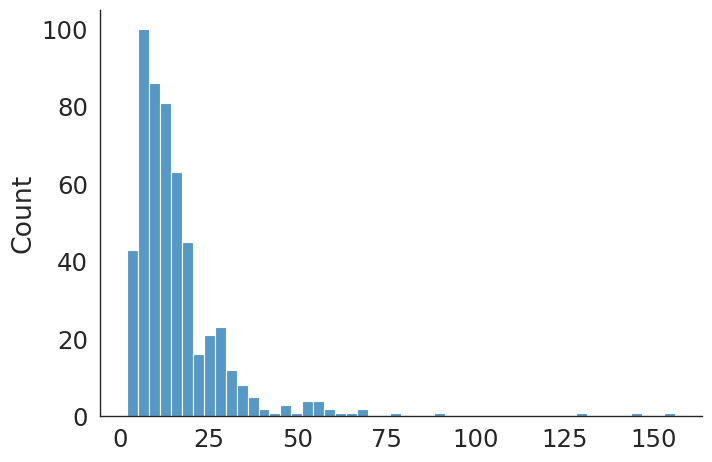
delay_los_pdv = pd.Series(delayed_los_v)
print("delay_los", delay_los_pdv.describe())
primary_knee_pdv = pd.Series(primary_knee_v)
print("primary_knee", primary_knee_pdv.describe())
uni_knee_pdv = pd.Series(uni_knee_v)
print("uni_knee", uni_knee_pdv.describe())
revise_knee_pdv = pd.Series(revise_knee_v)
print("revise_knee", revise_knee_pdv.describe())
primary_hip_pdv = pd.Series(primary_hip_v)
print("primary_hip", primary_hip_pdv.describe())
revise_hip_pdv = pd.Series(revise_hip_v)
print("revise_hip", revise_hip_pdv.describe())
delay_los count 529.000000
mean 16.521739
std 15.153132
min 2.000000
25% 8.000000
50% 13.000000
75% 19.000000
max 156.000000
dtype: float64
primary_knee count 2150.000000
mean 4.651163
std 2.828129
min 0.000000
25% 3.000000
50% 4.000000
75% 6.000000
max 42.000000
dtype: float64
uni_knee count 668.000000
mean 2.914671
std 2.136334
min 0.000000
25% 2.000000
50% 3.000000
75% 4.000000
max 16.000000
dtype: float64
revise_knee count 340.000000
mean 7.194118
std 7.598554
min 0.000000
25% 3.000000
50% 5.000000
75% 9.000000
max 63.000000
dtype: float64
primary_hip count 2820.000000
mean 4.433333
std 2.949526
min 0.000000
25% 3.000000
50% 4.000000
75% 5.000000
max 44.000000
dtype: float64
revise_hip count 406.000000
mean 6.908867
std 6.965812
min 0.000000
25% 3.000000
50% 5.000000
75% 8.000000
max 86.000000
dtype: float64
10. Convert to NumPy arrays for use in Fitter#
delay_los_npv = np.asarray(delayed_los_v)
primary_knee_npv = np.asarray(primary_knee_v)
uni_knee_npv = np.asarray(uni_knee_v)
revise_knee_npv = np.asarray(revise_knee_v)
primary_hip_npv = np.asarray(primary_hip_v)
revise_hip_npv = np.asarray(revise_hip_v)
11. Fit distributions#
get_distributions()
['_fit',
'alpha',
'anglit',
'arcsine',
'argus',
'beta',
'betaprime',
'bradford',
'burr',
'burr12',
'cauchy',
'chi',
'chi2',
'cosine',
'crystalball',
'dgamma',
'dweibull',
'erlang',
'expon',
'exponnorm',
'exponpow',
'exponweib',
'f',
'fatiguelife',
'fisk',
'foldcauchy',
'foldnorm',
'gamma',
'gausshyper',
'genexpon',
'genextreme',
'gengamma',
'genhalflogistic',
'genhyperbolic',
'geninvgauss',
'genlogistic',
'gennorm',
'genpareto',
'gibrat',
'gilbrat',
'gompertz',
'gumbel_l',
'gumbel_r',
'halfcauchy',
'halfgennorm',
'halflogistic',
'halfnorm',
'hypsecant',
'invgamma',
'invgauss',
'invweibull',
'johnsonsb',
'johnsonsu',
'kappa3',
'kappa4',
'ksone',
'kstwo',
'kstwobign',
'laplace',
'laplace_asymmetric',
'levy',
'levy_l',
'levy_stable',
'loggamma',
'logistic',
'loglaplace',
'lognorm',
'loguniform',
'lomax',
'maxwell',
'mielke',
'moyal',
'nakagami',
'ncf',
'nct',
'ncx2',
'norm',
'norminvgauss',
'pareto',
'pearson3',
'powerlaw',
'powerlognorm',
'powernorm',
'rayleigh',
'rdist',
'recipinvgauss',
'reciprocal',
'rice',
'rv_continuous',
'rv_histogram',
'semicircular',
'skewcauchy',
'skewnorm',
'studentized_range',
't',
'trapezoid',
'trapz',
'triang',
'truncexpon',
'truncnorm',
'truncweibull_min',
'tukeylambda',
'uniform',
'vonmises',
'vonmises_line',
'wald',
'weibull_max',
'weibull_min',
'wrapcauchy']
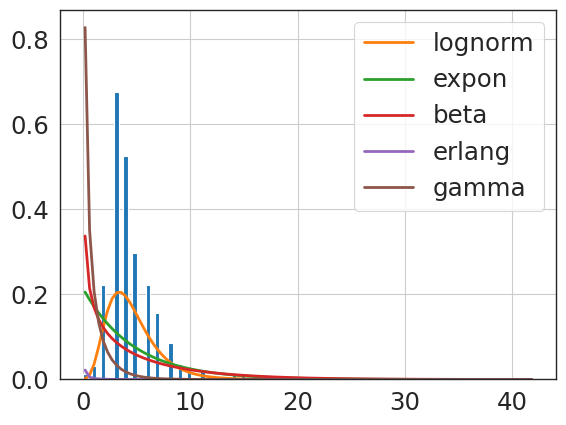
f = Fitter(delay_los_npv,
distributions=['gamma',
'lognorm',
'beta',
'expon',
'erlang'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 5 distributions: 100%|████████████████████| 5/5 [00:00<00:00, 27.36it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 0.004644 1608.122559 -6140.474568 inf 0.051474
beta 0.005357 1924.655190 -6058.553177 inf 0.087312
erlang 0.005441 1895.761508 -6056.655095 inf 0.088753
expon 0.009436 1599.608397 -5771.632045 inf 0.162747
gamma 0.035865 6380.344444 -5059.053541 inf 0.921926
ks_pvalue
lognorm 1.170505e-01
beta 5.866521e-04
erlang 4.477466e-04
expon 1.037166e-12
gamma 0.000000e+00
{'lognorm': {'s': 0.6977106156387711,
'loc': 0.46203959259225885,
'scale': 12.381893465319441}}
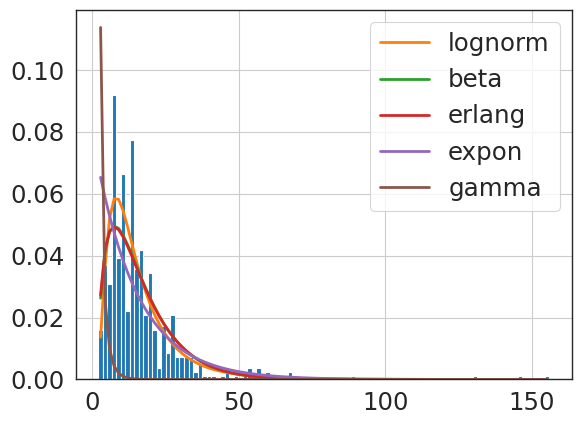
f = Fitter(primary_knee_npv,
distributions=['gamma',
'lognorm',
'beta',
'expon',
'erlang'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 5 distributions: 100%|████████████████████| 5/5 [00:00<00:00, 21.14it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 0.578484 1860.575987 -17651.201383 inf 0.151704
expon 0.792283 1214.423450 -16982.682686 inf 0.363244
beta 0.904699 1153.425425 -16682.064683 inf 0.427036
erlang 0.957660 4442.059841 -16567.424259 inf 0.992310
gamma 1.696721 2924.646338 -15337.709648 inf 0.872199
ks_pvalue
lognorm 1.150099e-43
expon 1.064843e-254
beta 0.000000e+00
erlang 0.000000e+00
gamma 0.000000e+00
{'lognorm': {'s': 0.4054742453283533,
'loc': -1.0156970361318063,
'scale': 5.187968679570998}}
f = Fitter(uni_knee_pdv,
distributions=['gamma',
'lognorm',
'beta',
'expon',
'erlang'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 5 distributions: 100%|████████████████████| 5/5 [00:00<00:00, 31.50it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 6.349747 924.541981 -3090.610424 inf 0.171962
gamma 6.363627 945.514562 -3089.151847 inf 0.167334
beta 6.373490 921.072492 -3081.613030 inf 0.168304
expon 6.603280 766.898464 -3070.961471 inf 0.271955
erlang 7.357514 3288.577391 -2992.209312 inf 0.919689
ks_pvalue
lognorm 9.648984e-18
gamma 8.102790e-17
beta 5.212017e-17
expon 3.872916e-44
erlang 0.000000e+00
{'lognorm': {'s': 0.3803355622865142,
'loc': -2.1623715729774657,
'scale': 4.71533144760631}}
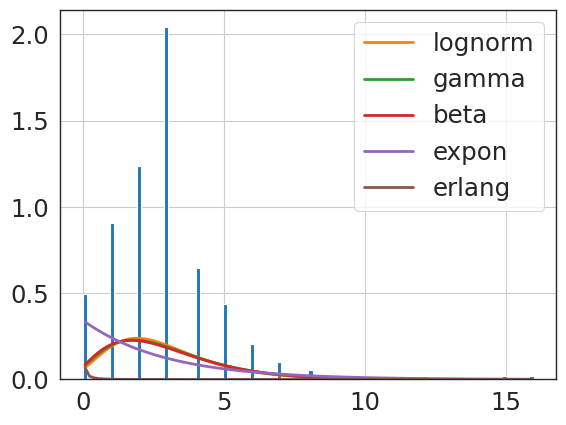
f = Fitter(revise_knee_npv,
distributions=['gamma',
'lognorm',
'beta',
'expon',
'erlang'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 5 distributions: 100%|████████████████████| 5/5 [00:00<00:00, 25.81it/s]
sumsquare_error aic bic kl_div ks_statistic \
expon 0.068790 1274.368194 -2880.262979 inf 0.123337
beta 0.100426 1301.742494 -2739.959169 inf 0.310421
lognorm 0.118113 1229.635547 -2690.633827 inf 0.366558
erlang 0.170774 5154.728107 -2565.275127 inf 0.895442
gamma 0.398495 4032.310584 -2277.175334 inf 0.699071
ks_pvalue
expon 5.759763e-05
beta 1.342060e-29
lognorm 1.816040e-41
erlang 0.000000e+00
gamma 5.534748e-167
{'expon': {'loc': 0.0, 'scale': 7.194117647058824}}
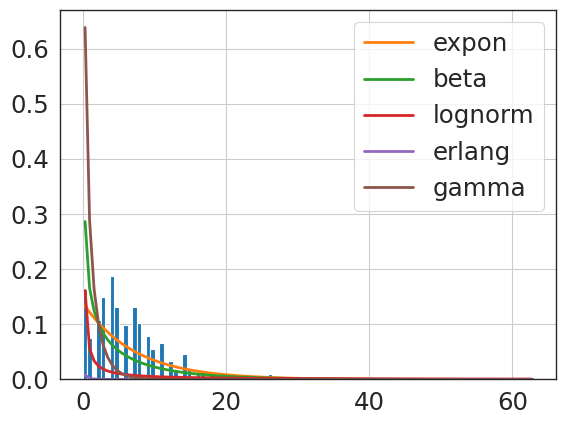
f = Fitter(primary_hip_npv,
distributions=['gamma',
'lognorm',
'beta',
'expon',
'erlang'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 5 distributions: 100%|████████████████████| 5/5 [00:00<00:00, 21.66it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 0.509158 1963.042781 -24283.125652 inf 0.166587
beta 0.516464 2432.283680 -24235.005142 inf 0.168023
expon 0.654193 1294.311552 -23584.255723 inf 0.334257
gamma 0.686329 1119.561080 -23441.078834 inf 0.325523
erlang 0.828594 4614.364362 -22909.864887 inf 0.962388
ks_pvalue
lognorm 7.205752e-69
beta 4.630348e-70
expon 2.091397e-281
gamma 1.536661e-266
erlang 0.000000e+00
{'lognorm': {'s': 0.3615166592899655,
'loc': -2.4363376840086204,
'scale': 6.4116655364890836}}
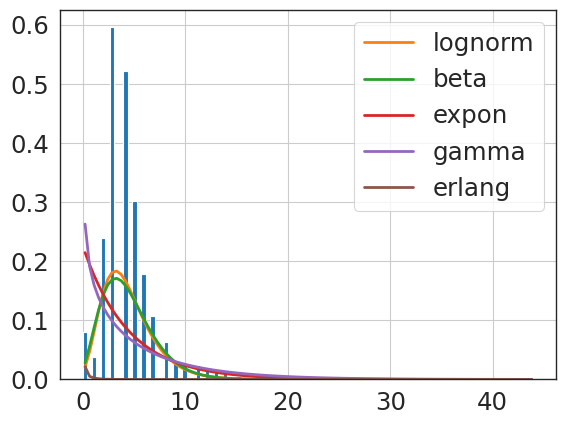
f = Fitter(revise_hip_npv,
distributions=['gamma',
'lognorm',
'beta',
'expon',
'erlang'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 5 distributions: 100%|████████████████████| 5/5 [00:00<00:00, 21.51it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 0.022496 1733.019612 -3961.087365 inf 0.107411
beta 0.033304 2128.183021 -3795.802654 inf 0.142546
expon 0.059486 1635.338315 -3572.307506 inf 0.241396
erlang 0.130983 5711.381127 -3245.831210 inf 0.983865
gamma 0.138877 3996.315953 -3222.070957 inf 0.938027
ks_pvalue
lognorm 1.559684e-04
beta 1.162057e-07
expon 2.614308e-21
erlang 0.000000e+00
gamma 0.000000e+00
{'lognorm': {'s': 0.6152292643165856,
'loc': -0.934387442319201,
'scale': 6.350152207311825}}
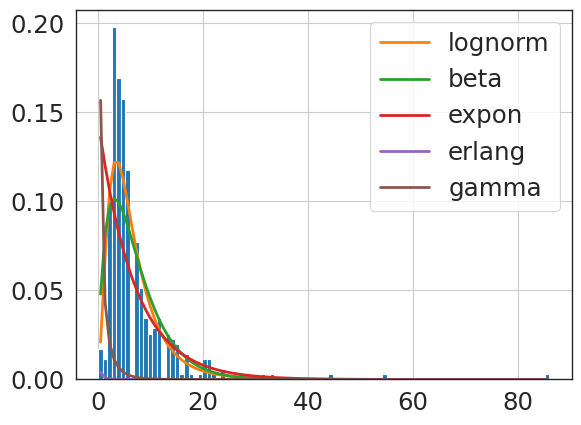
f.get_best(method = 'sumsquare_error')
{'lognorm': {'s': 0.6152292643165856,
'loc': -0.934387442319201,
'scale': 6.350152207311825}}
12. Clip arrays - optional truncation of los data#
delay_los_npv_clip = np.asarray(delayed_los_v) # leave as is
primary_knee_npv_clip = np.clip(np.asarray(primary_knee_v), 0, 30)
uni_knee_npv_clip = np.clip(np.asarray(uni_knee_v), 0, 30)
revise_knee_npv_clip = np.clip(np.asarray(revise_knee_v), 0, 30)
primary_hip_npv_clip = np.clip(np.asarray(primary_hip_v), 0, 30)
revise_hip_npv_clip = np.clip(np.asarray(revise_hip_v), 0, 30)
12.1 Fit clipped data#
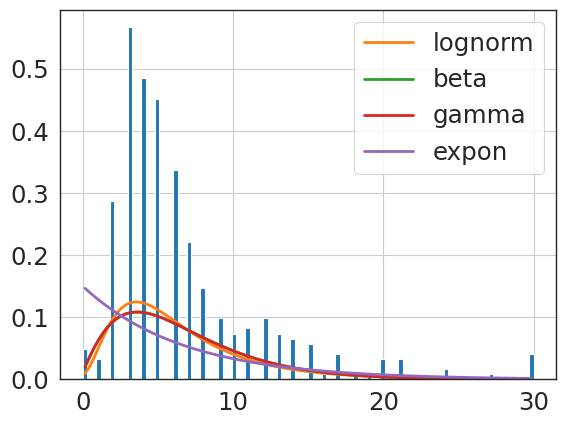
f = Fitter(primary_knee_npv_clip,
distributions=['gamma',
'lognorm',
'beta',
'expon'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 4 distributions: 100%|████████████████████| 4/4 [00:00<00:00, 28.55it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 1.352282 1383.035323 -15825.554283 inf 0.151034
gamma 1.369003 1633.491636 -15799.132769 inf 0.167515
beta 1.371934 1592.110062 -15786.860642 inf 0.166926
expon 1.651798 957.270922 -15403.074367 inf 0.363888
ks_pvalue
lognorm 2.775736e-43
gamma 3.330412e-53
beta 7.852058e-53
expon 1.245667e-255
{'lognorm': {'s': 0.40225541615533966,
'loc': -1.0399482363715324,
'scale': 5.212995574296882}}
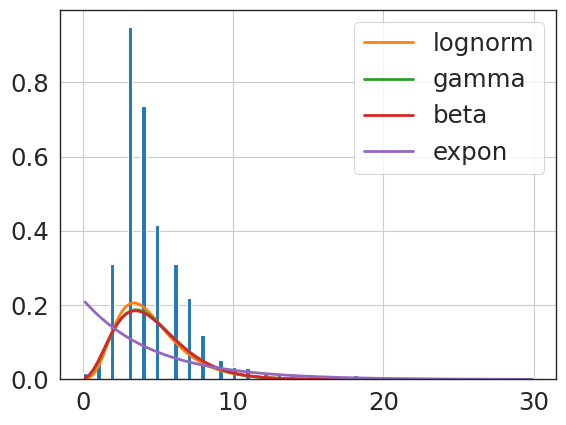
f = Fitter(uni_knee_npv_clip,
distributions=['gamma',
'lognorm',
'beta',
'expon'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 4 distributions: 100%|████████████████████| 4/4 [00:00<00:00, 43.09it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 6.349747 924.541981 -3090.610424 inf 0.171962
gamma 6.363627 945.514562 -3089.151847 inf 0.167334
beta 6.373490 921.072492 -3081.613030 inf 0.168304
expon 6.603280 766.898464 -3070.961471 inf 0.271955
ks_pvalue
lognorm 9.648984e-18
gamma 8.102790e-17
beta 5.212017e-17
expon 3.872916e-44
{'lognorm': {'s': 0.3803355622865142,
'loc': -2.1623715729774657,
'scale': 4.71533144760631}}
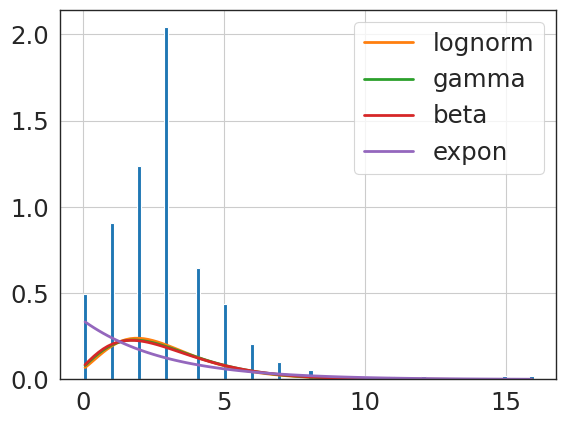
f = Fitter(revise_knee_npv_clip,
distributions=['gamma',
'lognorm',
'beta',
'expon'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 4 distributions: 100%|████████████████████| 4/4 [00:00<00:00, 29.05it/s]
sumsquare_error aic bic kl_div ks_statistic \
expon 0.553174 824.389317 -2171.491717 inf 0.134110
beta 0.567029 862.802255 -2151.423185 inf 0.246918
lognorm 0.614330 1100.527541 -2130.010433 inf 0.399620
gamma 0.635206 904.170953 -2118.648631 inf 0.369219
ks_pvalue
expon 8.562804e-06
beta 9.612934e-19
lognorm 1.649474e-49
gamma 4.379593e-42
{'expon': {'loc': 0.0, 'scale': 6.920588235294118}}
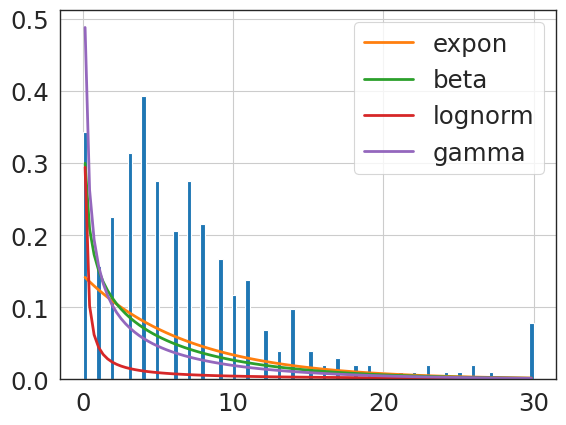
f = Fitter(primary_hip_npv_clip,
distributions=['gamma',
'lognorm',
'beta',
'expon'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 4 distributions: 100%|████████████████████| 4/4 [00:00<00:00, 29.92it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 1.309768 1377.265202 -21618.657207 inf 0.165827
gamma 1.321269 1546.157021 -21594.003778 inf 0.166356
beta 1.324767 1505.611887 -21578.602914 inf 0.169617
expon 1.537796 979.324423 -21173.990919 inf 0.334836
ks_pvalue
lognorm 3.051803e-68
gamma 1.117626e-68
beta 2.135810e-71
expon 2.091822e-282
{'lognorm': {'s': 0.35741083072060453,
'loc': -2.489064861474697,
'scale': 6.466619066862258}}
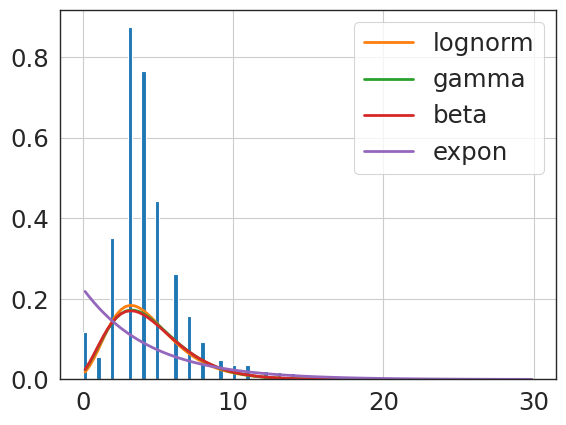
f = Fitter(revise_hip_npv_clip,
distributions=['gamma',
'lognorm',
'beta',
'expon'
]);
f.fit()
print(f.summary())
f.get_best(method = 'sumsquare_error')
Fitting 4 distributions: 100%|████████████████████| 4/4 [00:00<00:00, 44.59it/s]
sumsquare_error aic bic kl_div ks_statistic \
lognorm 0.780455 903.666307 -2521.198708 inf 0.106709
beta 0.796737 919.212449 -2506.809528 inf 0.132526
gamma 0.796812 916.728918 -2512.777811 inf 0.132578
expon 0.885004 833.485595 -2476.164953 inf 0.251605
ks_pvalue
lognorm 1.764127e-04
beta 1.116592e-06
gamma 1.104079e-06
expon 3.889626e-23
{'lognorm': {'s': 0.5796389432896831,
'loc': -1.102767801493329,
'scale': 6.517280834355917}}
12.2 Summarise clipped data#
delay_los_pdv = pd.Series(delay_los_npv_clip)
print("delay_los", delay_los_pdv.describe())
primary_knee_pdv = pd.Series(primary_knee_npv_clip)
print("primary_knee", primary_knee_pdv.describe())
uni_knee_pdv = pd.Series(uni_knee_npv_clip)
print("uni_knee", uni_knee_pdv.describe())
revise_knee_pdv = pd.Series(revise_knee_npv_clip)
print("revise_knee", revise_knee_pdv.describe())
primary_hip_pdv = pd.Series(primary_hip_npv_clip)
print("primary_hip", primary_hip_pdv.describe())
revise_hip_pdv = pd.Series(revise_hip_npv_clip)
print("revise_hip", revise_hip_pdv.describe())
delay_los count 529.000000
mean 16.521739
std 15.153132
min 2.000000
25% 8.000000
50% 13.000000
75% 19.000000
max 156.000000
dtype: float64
primary_knee count 2150.000000
mean 4.642326
std 2.731335
min 0.000000
25% 3.000000
50% 4.000000
75% 6.000000
max 30.000000
dtype: float64
uni_knee count 668.000000
mean 2.914671
std 2.136334
min 0.000000
25% 2.000000
50% 3.000000
75% 4.000000
max 16.000000
dtype: float64
revise_knee count 340.000000
mean 6.920588
std 6.291085
min 0.000000
25% 3.000000
50% 5.000000
75% 9.000000
max 30.000000
dtype: float64
primary_hip count 2820.000000
mean 4.425887
std 2.869115
min 0.000000
25% 3.000000
50% 4.000000
75% 5.000000
max 30.000000
dtype: float64
revise_hip count 406.000000
mean 6.665025
std 5.232482
min 0.000000
25% 3.000000
50% 5.000000
75% 8.000000
max 30.000000
dtype: float64

